Advertisement |
 |
nature.com webcasts
Springer Nature presents a custom webcast on: 3D High-Content Screening of Organoids for Drug Discovery.
Register for FREE
Date: Wednesday 24th May
Time: 8AM PDT | 11AM EDT | 4PM BST | 5PM CEST
Sponsored by: Perkin Elmer
www.PerkinElmer.com/cellularimaging |  | | |
 |
 |
TABLE OF CONTENTS
|
June 2017 Volume 13, Issue 6 |
 |  |  |
 | 
 Editorial Editorial
 Commentaries Commentaries
 Research Highlights Research Highlights
 News and Views News and Views
 Perspectives Perspectives
 Review Review
 Brief Communications Brief Communications
 Articles Articles
 Errata Errata
 Corrigendum Corrigendum
| |
 |
|
 |
 |
Advertisement |
 |
| The 1st Symposium on Nucleic Acid Modifications will bring together key players in the previously segregated fields of RNA and DNA modifications, which are now merging to create a brand new interdisciplinary research field that includes chemical biology, structural biology, bioinformatics, biophysics and cell biology, developmental biology and epigenetics. The symposium will outline the major developments in this rapidly developing field, and includes talks from leaders in this exciting area of study. www.imb.de/2017nucmod |  | | |
 |
| |
Focus |  Top Top |
 |
 |
 | Focus on Chemical approaches in developmental biology
 The development of model organisms such as zebrafish and worms progresses from a single cell to tissues and organs. In this issue, a collection of articles highlights advances and opportunities at the intersection of developmental and chemical biology. The development of model organisms such as zebrafish and worms progresses from a single cell to tissues and organs. In this issue, a collection of articles highlights advances and opportunities at the intersection of developmental and chemical biology.
Focus on Chemical approaches in developmental biology
|
 |
Editorial |  Top Top |
 |
 |
 |
Shaping embryonic development p559
doi:10.1038/nchembio.2403
The growing intersection between chemical tools and principles and developmental biology is providing new insights into the molecular-level details of developmental processes.
|
 |
Commentaries |  Top Top |
 |
 |
 |
Small-molecule phenotypic screening with stem cells pp560 - 563
Andrei Ursu, Hans R Scholer and Herbert Waldmann
doi:10.1038/nchembio.2383
To fully leverage the potential of human-induced pluripotent stem cells (hiPSCs), improved and standardized reprogramming methods and large-scale collections of hiPSC lines are needed, and the stem cell community must embrace chemical biology methodology for target identification and validation.
|
 |
 |
 |
Unraveling cell-to-cell signaling networks with chemical biology pp564 - 568
Zev J Gartner, Jennifer A Prescher and Luke D Lavis
doi:10.1038/nchembio.2391
Cell-to-cell signaling networks, although poorly understood, guide tissue development, regulate tissue function and may become dysregulated in disease. Chemical biologists can develop the next generation of tools to untangle these complex and dynamic networks of interacting cells.
|
 |
Research Highlights |  Top Top |
 |
 |
 |
Synthetic biology: Return to sender | Enzymology: Radical ring resizing | Infectious disease: A lethal sugar fix | Plant development: Get lit on steroids
|
News and Views |  Top Top |
 |
 |
 |
| |
 |
Perspectives |  Top Top |
 |
 |
 |
Small-molecule pheromones and hormones controlling nematode development pp577 - 586
Rebecca A Butcher
doi:10.1038/nchembio.2356
|
 |
 |
 |
Illuminating developmental biology through photochemistry pp587 - 598
Lukasz Kowalik and James K Chen
doi:10.1038/nchembio.2369
|
 |
Review |  Top Top |
 |
 |
 |
The perception of strigolactones in vascular plants pp599 - 606
Shelley Lumba, Duncan Holbrook-Smith and Peter McCourt
doi:10.1038/nchembio.2340
|
 |
Brief Communications |  Top Top |
 |
 |
 |
|
 |
 |
 |
Structural and functional insight into human O-GlcNAcase pp610 - 612
Christian Roth, Sherry Chan, Wendy A Offen, Glyn R Hemsworth, Lianne I Willems et al.
doi:10.1038/nchembio.2358
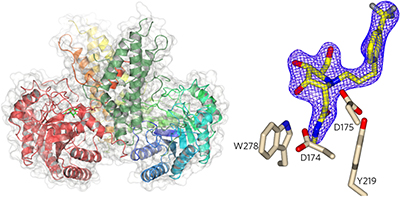
Crystal structures of human O-GlcNAc hydrolase (hOGA) fragments show that hOGA's dimeric structure is organized by swapping of an α-helical element and reveal features of inhibitor binding to the catalytic domain.
|
|
|
 |
 |
 |
|
 |
Articles |  Top Top |
 |
 |
 |
Chemical screening identifies ATM as a target for alleviating senescence pp616 - 623
Hyun Tae Kang, Joon Tae Park, Kobong Choi, Yongsub Kim, Hyo Jei Claudia Choi et al.
doi:10.1038/nchembio.2342
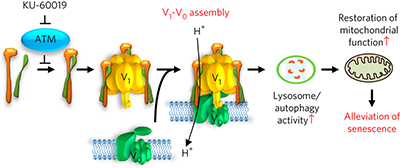
Ataxia telangiectasia mutated (ATM) directly interacts with and phosphorylates the V-ATPase V1 subunit ATP6V1G1, thereby decreasing V1-V0 assembly in the V-ATPase. Attenuation of ATM activity results in lysosomal pH acidification, recovery of autophagy and alleviation of senescence.
|
|
|
 |
 |
 |
|
 |
 |
 |
Near-infrared optogenetic pair for protein regulation and spectral multiplexing pp633 - 639
Taras A Redchuk, Evgeniya S Omelina, Konstantin G Chernov and Vladislav V Verkhusha
doi:10.1038/nchembio.2343

The engineering of Q-PAS1, a single-domain variant of PpsR2, led to an optimized optogenetic system based on the Q-PAS1-BphP1 interaction, which was applied to the regulation of transcription, epigenetic state and protein localization by near-infrared light.
|
|
|
 |
 |
 |
AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet pp640 - 646
Dukas Jurenas, Sneha Chatterjee, Albert Konijnenberg, Frank Sobott, Louis Droogmans et al.
doi:10.1038/nchembio.2346
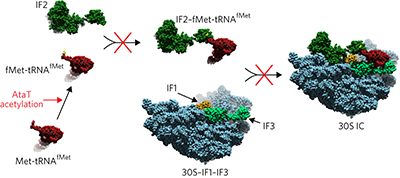
Characterization of an enterohaemorrhagic E. coli toxin-antitoxin system reveals that the toxin AtaT specifically acetylates Met-tRNAfMet at the methionyl amine, making it incompetent for translation initiation, which inhibits translation.
|
|
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
Global survey of the immunomodulatory potential of common drugs pp681 - 690
Gregory I Vladimer, Berend Snijder, Nikolaus Krall, Johannes W Bigenzahn, Kilian V M Huber et al.
doi:10.1038/nchembio.2360
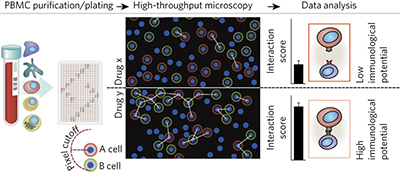
A high-content screening platform that measures the immunological potential of small-molecule and biologic drugs by computationally determining changes in the physical interactions among peripheral mononuclear leukocytes revealed known immunomodulators and also approved drugs as regulators of unexpected targets, including MST1R.
|
|
|
 |
Corrigendum |  Top Top |
 |
 |
 |
Corrigendum: Full antagonism of the estrogen receptor without a prototypical ligand side chain p691
Sathish Srinivasan, Jerome C Nwachukwu, Nelson E Bruno, Venkatasubramanian Dharmarajan, Devrishi Goswami et al.
doi:10.1038/nchembio0617-691c
|
 |
Errata |  Top Top |
 |
 |
 |
Erratum: A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation p691
Kai Papenfort, Justin E Silpe, Kelsey R Schramma, Jian-Ping Cong, Mohammad R Seyedsayamdost et al.
doi:10.1038/nchembio0617-691a
|
 |
 |
 |
Erratum: Full antagonism of the estrogen receptor without a prototypical ligand side chain p691
Sathish Srinivasan, Jerome C Nwachukwu, Nelson E Bruno, Venkatasubramanian Dharmarajan, Devrishi Goswami et al.
doi:10.1038/nchembio0617-691b
|
 |
 Top Top |
 |
 |
 |  |  |  |  |  | Natureevents is a fully searchable, multi-disciplinary database designed to maximise exposure for events organisers. The contents of the Natureevents Directory are now live. The digital version is available here.
Find the latest scientific conferences, courses, meetings and symposia on natureevents.com. For event advertising opportunities across the Nature Publishing Group portfolio please contact natureevents@nature.com |  |  |  |  |  |
|
 |


No comments:
Post a Comment