TABLE OF CONTENTS
|
April 2017 Volume 13, Issue 4 |
 |  |  |
 |  Commentary Commentary
 Research Highlights Research Highlights
 News and Views News and Views
 Review Review
 Brief Communications Brief Communications
 Articles Articles
| |
 |
|
 |
 |
| Advertisement |
 |
| |
 |
| |
Advertisement |
 |
nature.com webcasts
Join us for our upcoming webcast followed by live Q&A:
NEW INNOVATIONS IN DRUG DELIVERY TECHNOLOGY
Presented by BioPharma Dealmakers
Date: Wednesday, March 29, 2017
Time: 8AM PDT | 11AM EDT | 4PM BST | 5PM CEST
REGISTER FOR FREE
Sponsored by: Corium International | Portal Instruments Inc. | TissueGen Inc. | TrioxNano | | | |
 |
| |
Commentary |  Top Top |
 |
 |
 |
Simultaneous quantification of protein order and disorder pp339 - 342
Pietro Sormanni, Damiano Piovesan, Gabriella T Heller, Massimiliano Bonomi, Predrag Kukic et al.
doi:10.1038/nchembio.2331
Nuclear magnetic resonance spectroscopy is transforming our views of proteins by revealing how their structures and dynamics are closely intertwined to underlie their functions and interactions. Compelling representations of proteins as statistical ensembles are uncovering the presence and biological relevance of conformationally heterogeneous states, thus gradually making it possible to go beyond the dichotomy between order and disorder through more quantitative descriptions that span the continuum between them.
|
 |
Research Highlights |  Top Top |
 |
 |
 |
Development: Marking the transition | Bioconjugation: Methionine's time to shine | Drug discovery: Uncoupling coupled transport | Imaging: Luciferase matchmaker
|
News and Views |  Top Top |
 |
 |
 |
| |
 |
Review |  Top Top |
 |
 |
 |
Versatile modes of cellular regulation via cyclic dinucleotides pp350 - 359
Petya Violinova Krasteva and Holger Sondermann
doi:10.1038/nchembio.2337
A review of the roles of cyclic dinucleotides (CDNs) in signaling systems including transcription, ion transport, bacterial secretion and eukaryotic immune responses, highlighting the diverse binding modes of CDNs by target proteins and functional insights gained from structural studies.
|
 |
Brief Communications |  Top Top |
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
Ligand-promoted protein folding by biased kinetic partitioning pp369 - 371
Karan S Hingorani, Matthew C Metcalf, Derrick T Deming, Scott C Garman, Evan T Powers et al.
doi:10.1038/nchembio.2303
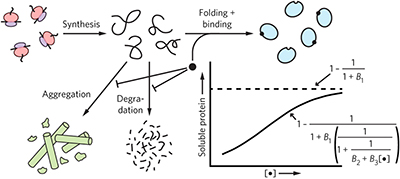
Pharmacological chaperones improve folding of destabilized Escherichia coli dihydrofolate reductase (DHFR) and human disease-linked α-galactosidase A (α-GAL) by biasing the kinetic partitioning between folding, aggregation, and degradation. Chaperoning spares DHFR from aggregation and α-GAL from degradation.
See also: News and Views by Hatters
|
|
|
 |
| Advertisement |
 |
- Reach a potential audience of 10.8 million users* via nature.com
- Be seen first by candidates — your position will appear at the top of related searches for 60 days and be displayed prominently on the Naturejobs home page as Job of the Week.
- Plus appear as 'Featured' in search results
| | | |
 |
| |
Articles |  Top Top |
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
A water-mediated allosteric network governs activation of Aurora kinase A pp402 - 408
Soreen Cyphers, Emily F Ruff, Julie M Behr, John D Chodera and Nicholas M Levinson
doi:10.1038/nchembio.2296
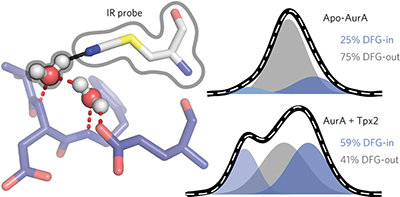
Spectroscopic studies of allosteric activation of Aurora A kinase using a site-specific infrared probe combined with FRET analysis and molecular dynamics simulations reveals a water-mediated hydrogen bond network in the active site that regulates Aurora A activity.
|
|
|
 |
 |
 |
A tight tunable range for Ni(II) sensing and buffering in cells pp409 - 414
Andrew W Foster, Rafael Pernil, Carl J Patterson, Andrew J P Scott, Lars-Olof Palsson et al.
doi:10.1038/nchembio.2310
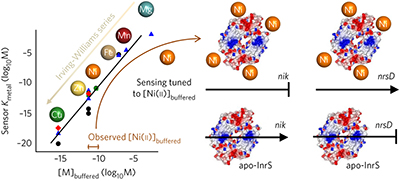
The Ni(ii) affinity of Ni(ii) sensor InrS is attuned to buffered Ni(ii) concentrations, explaining why these two parameters co-vary for different metals over many orders of magnitude.
|
|
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
Evolution of a split RNA polymerase as a versatile biosensor platform pp432 - 438
Jinyue Pu, Julia Zinkus-Boltz and Bryan C Dickinson
doi:10.1038/nchembio.2299
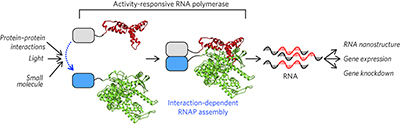
Design of a proximity-dependent split RNA polymerase system and its optimization by phage-assisted continuous evolution (PACE) enabled the development of a family of activity-dependent split RNA polymerase biosensors regulated by small molecules or light.
|
|
|
 |
 |
 |
The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme pp439 - 445
Jamie L Bingaman, Sixue Zhang, David R Stevens, Neela H Yennawar, Sharon Hammes-Schiffer et al.
doi:10.1038/nchembio.2300
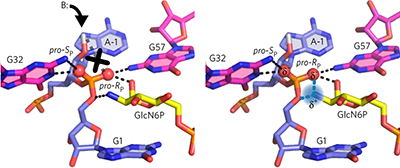
Experimental work and computational modeling together reveal a suite of catalytic roles of the GlcN6P cofactor in the glmS ribozyme, including activation of the nucleophile, electrostatic stabilization, and alignment of the active site.
|
|
|
 |
 |
 |
|
 |
 |  |  |  |  |  | Natureevents is a fully searchable, multi-disciplinary database designed to maximise exposure for events organisers. The contents of the Natureevents Directory are now live. The digital version is available here.
Find the latest scientific conferences, courses, meetings and symposia on natureevents.com. For event advertising opportunities across the Nature Publishing Group portfolio please contact natureevents@nature.com |  |  |  |  |  |
|
 |


No comments:
Post a Comment